Google is taking another major swing at genomics, and this time it’s targeting the genetic chaos of cancer. The company just released Google DeepSomatic, an open-source AI model designed to pinpoint the specific DNA mutations that drive tumor growth with what it claims is unprecedented accuracy. Building on the foundation of its well-regarded DeepVariant tool, this new model tackles the notoriously messy world of somatic mutations—the genetic errors a tumor acquires as it grows.
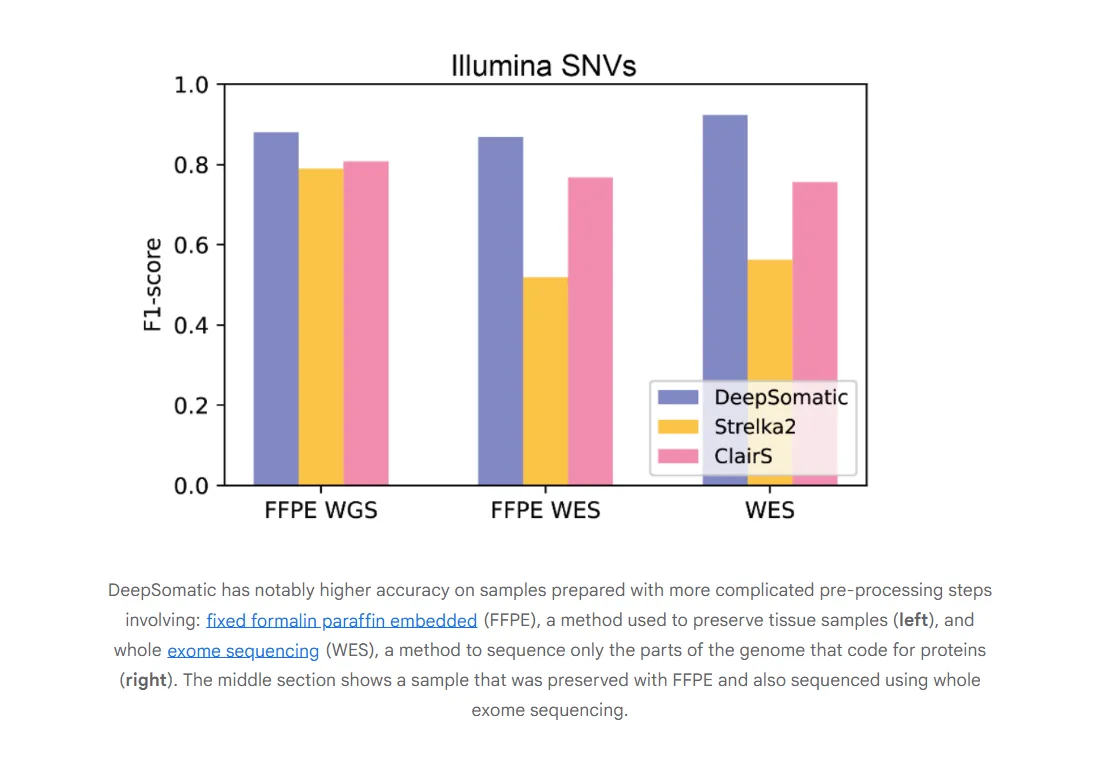
Developed in a joint effort with the UC Santa Cruz Genomics Institute and Children’s Mercy Hospital, DeepSomatic isn’t just an incremental update. It fundamentally changes how mutation detection works. The model transforms raw, noisy DNA sequencing data into image-like files, then uses a convolutional neural network (CNN) to visually distinguish between three things: harmless inherited DNA, critical cancer-causing mutations, and simple sequencing errors. This image-based approach allows the AI to find patterns even in the low-quality, fragmented DNA samples common in clinical settings, like tissue preserved in formaldehyde.
The goal here is to supercharge precision oncology—the practice of tailoring treatments to a tumor’s unique genetic fingerprint. By accurately identifying the mutations fueling a cancer, clinicians can choose more effective therapies. And by open-sourcing the model and its high-quality training data, Google is making a clear play to set a new industry standard, just as it did with DeepVariant for inherited diseases.
From Lab Bench to Clinical Benchmark
This isn't just a theoretical exercise. According to a paper published in *Nature Biotechnology*, Google DeepSomatic significantly outperforms existing bioinformatics tools. On standard Illumina sequencing data, it achieved a 90 percent F1-score for detecting tricky insertion and deletion mutations, a notable jump from the 80 percent managed by the next-best tool. The leap was even more dramatic on long-read PacBio data, where DeepSomatic’s accuracy more than doubled that of conventional methods, jumping from below 50 percent to over 80 percent.
Crucially, the model excels at "tumor-only" analysis. For blood cancers like leukemia, getting a clean sample of non-cancerous DNA is often impossible. DeepSomatic was trained to work around this, learning to isolate tumor-specific signals without a healthy baseline for comparison. This capability alone could unlock new research and diagnostic avenues that were previously out of reach.
The release includes the CASTLE reference dataset built with the National Cancer Institute, which was used to train the model across three different sequencing platforms. This cross-platform validation is key to eliminating platform-specific biases and creating a truly universal tool.
By packaging DeepSomatic with other tools like Severus for detecting larger genomic changes, Google and its partners are building a comprehensive, open-source toolkit for cancer genomics. It’s a strategic move to embed its AI deep within the research and clinical pipeline, bridging the gap between raw sequence data and actionable medical insight.