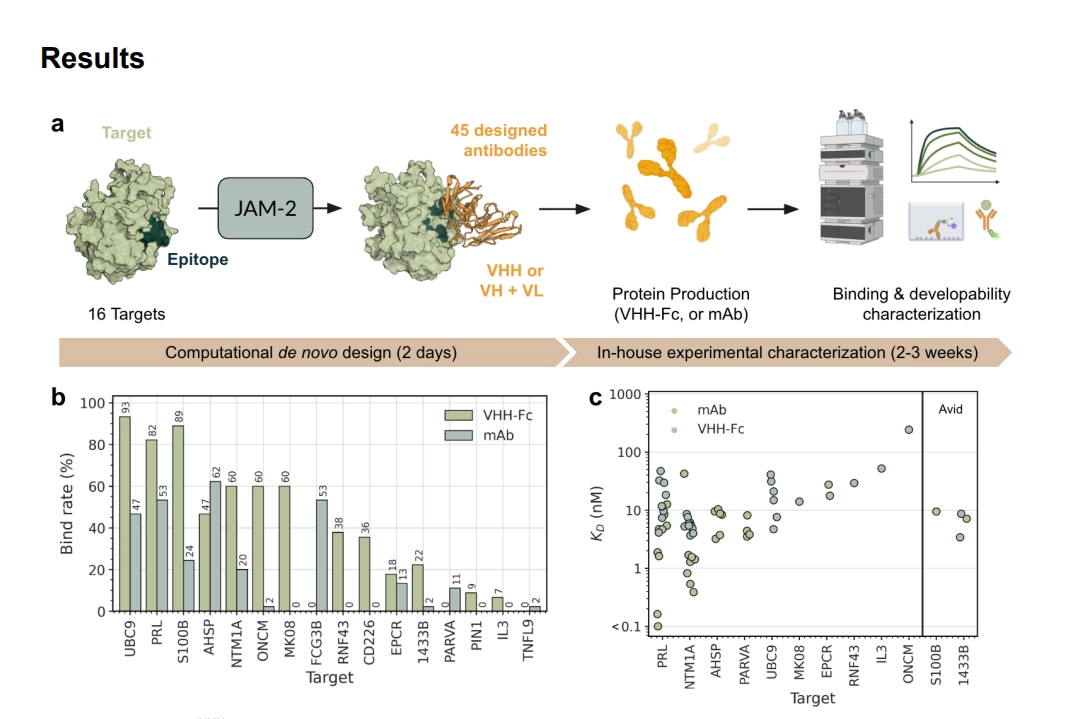
The era of relying on the messy, stochastic process of immunization for antibody discovery may be drawing to a close. Nabla Bio has unveiled JAM-2, a fully computational system for de novo protein design that is demonstrating performance metrics previously thought to be years away for AI-driven biology. This isn't just about generating more antibodies; it’s about generating better ones, faster, and targeting sites that traditional methods consistently miss.
The core breakthrough with JAM-2, detailed by Nabla Bio, is its ability to bypass the traditional bottlenecks of affinity maturation and developability screening. In testing against 16 structurally diverse, unseen targets, JAM-2 achieved a 100% hit rate across both VHH-Fc and full-length mAb formats. Crucially, half of these initial computer-generated binders already exhibited picomolar or low nanomolar affinity—a level that usually requires extensive lab optimization. The efficiency gain is stark: achieving these results required screening only dozens of designs computationally, contrasting sharply with the billions of variants churned out by phage display libraries.
Precision Control Over Functionality
Perhaps the most significant shift JAM-2 offers drug developers is the move from serendipity to intentional design. Traditional antibody discovery is plagued by "immunodominance," where the immune system or screening libraries only find binders against the most exposed, accessible parts of a target protein. This often means missing critical functional sites necessary for therapeutic effect.
JAM-2 allows users to specify the exact epitope—the binding patch—on the target antigen. Nabla Bio demonstrated success rates between 30% and 70% in generating binders for user-defined patches on half of the tested targets. This level of precision means researchers can now design antibodies to specifically induce agonism or antagonism, or deliberately avoid regions linked to off-target effects, fundamentally changing the strategic approach to target validation.
This capability proved vital when tackling G-protein-coupled receptors (GPCRs), a notoriously difficult class of membrane proteins. JAM-2 successfully designed antibodies that bind directly to the orthosteric pockets of CXCR4 and CXCR7—the primary binding sites—while the proteins were still in their native cellular context. The resulting affinities (single-digit nanomolar) matched or exceeded those from lengthy immunization campaigns, suggesting a viable path to drugging targets previously deemed too complex for antibody intervention.
Furthermore, the system appears to have solved the common AI pitfall where functional designs are biophysically useless. Hundreds of JAM-2 outputs were subjected to industrial-grade developability profiling. A remarkable 57% of designs passed all four primary criteria (expression, stability, low aggregation, and low stickiness) simultaneously. The resulting sequences showed high human germline identity, suggesting low immunogenicity risk, with top candidates exhibiting profiles comparable to established drugs like Trastuzumab.
The implications for Big Pharma and biotech are clear: JAM-2 positions generative antibody design as a high-throughput, reliable engine capable of parallel discovery across vast target panels. By collapsing the timeline from design to purified, optimized protein from months or years down to weeks, Nabla Bio is signaling a major inflection point where computational prediction finally outpaces wet-lab screening in reliability and speed.